Candidate Sampling
参考:https://www.tensorflow.org/extras/candidate_sampling.pdf
Why Candidate Sampling
| “Exhaustive” training methods such as softmax and logistic regression require us to compute F(x, y) for every class y ∈ L for every training example. When | L | is very large, this can be prohibitively expensive. |
“Candidate Sampling” training methods involve constructing a training task in which for each training example $(x_i,T_i)$ , we only need to evaluate $F(x, y)$ for a small set of candidate classes $C_i\subset L$ . Typically, the set of candidates $C_i$ is the union of the target classes with a randomly chosen sample of (other) classes $S_i\subset L$ .
The random choice of $S_i$ may or may not depend on $x_i$ and/or $T_i$.
Algorithms
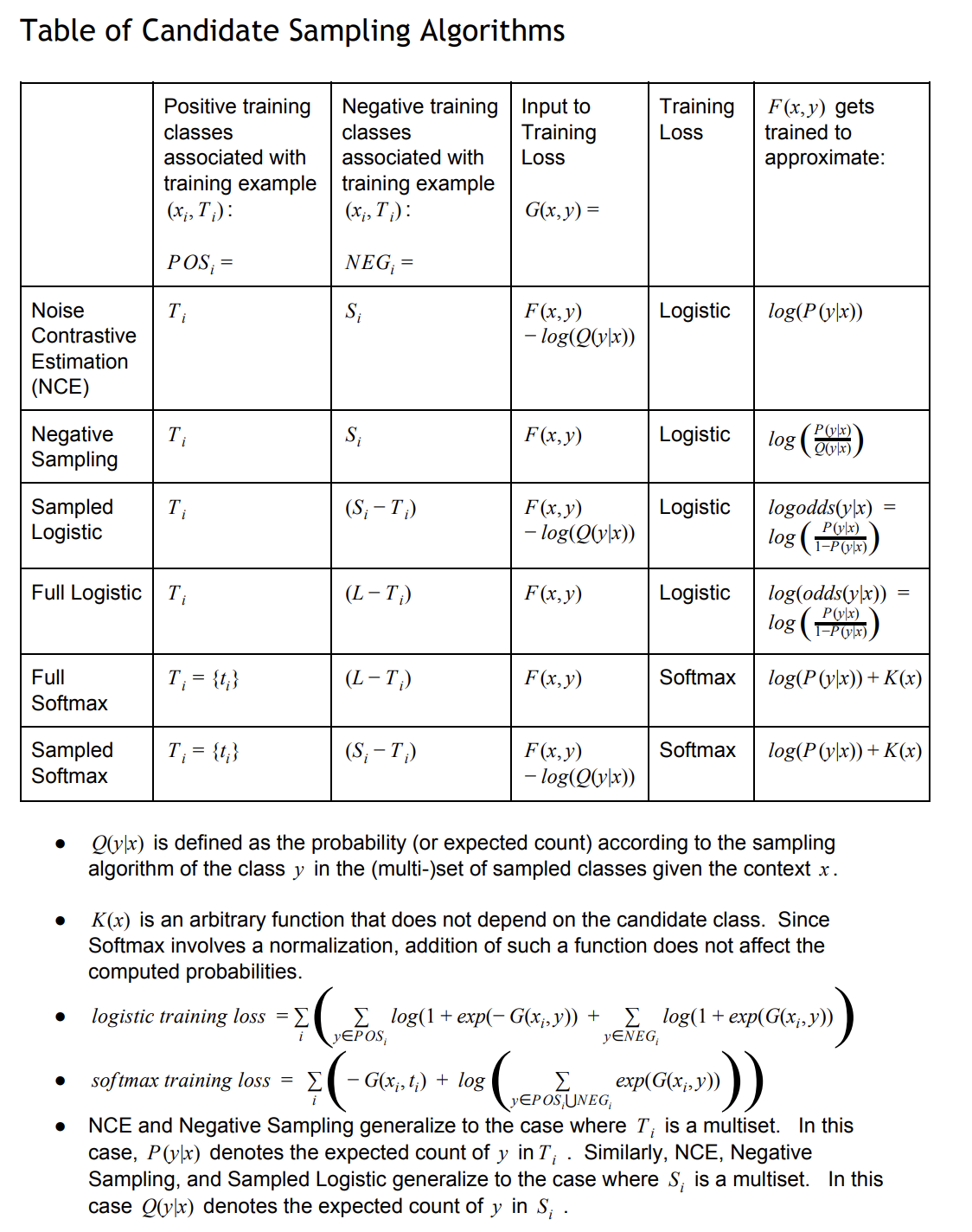
一些表的的解释:
-
$Q(y x)$是给定context $x$时,y(被采样到的集合里面的label)的概率或者expect count。如果采样的负样本都不重复,就是概率,否则,就是采样个数(expect count); -
最后一列表示这个algorithm要近似的目标,因为都是采样部分负样本,无法完全准确的计算每个label的分数,只能近似。比如NCE用$F(x,y)-log(Q(y x))$来近似$log(P(y x))$。
Candidate Sampling in TensorFlow
Tensorflow实现了两种常用于word2vec的loss,sampled softmax和NCE,这两种loss本身可以用于任意分类问题(single-label or multi-label)。两个方法的目标是在分类的数量太大时,简化softmax计算,采取估算的方法得到softmax的值。
Intuitive explanation
-
NCE loss的直观想法:把多分类问题转化成二分类。
之前计算softmax的时候class数量太大,NCE索性就把分类缩减为二分类问题。之前的问题是计算某个类的归一化概率是多少,二分类的问题是input和label正确匹配的概率是多少。二分类问题群众喜闻乐见,直接上logistic regression估算一下概率。
-
Sampled softmax则是只抽取一部分样本计算softmax。这个想法也很好理解,训练的时候我不需要特别精准的softmax归一化概率,我只需要一个粗略值做back propoagation就好了
此部分参考: https://www.zhihu.com/question/50043438/answer/254300443
Code View
首先我们来看看这两个loss的他们的核心代码_compute_sampled_logits:
def _compute_sampled_logits(weights,
biases,
labels,
inputs,
num_sampled,
num_classes,
num_true=1,
sampled_values=None,
subtract_log_q=True,
remove_accidental_hits=False,
partition_strategy="mod",
name=None,
seed=None):
"""Helper function for nce_loss and sampled_softmax_loss functions.
Computes sampled output training logits and labels suitable for implementing
e.g. noise-contrastive estimation (see nce_loss) or sampled softmax (see
sampled_softmax_loss).
Note: In the case where num_true > 1, we assign to each target class
the target probability 1 / num_true so that the target probabilities
sum to 1 per-example.
Args:
weights: A `Tensor` of shape `[num_classes, dim]`, or a list of `Tensor`
objects whose concatenation along dimension 0 has shape
`[num_classes, dim]`. The (possibly-partitioned) class embeddings.
biases: A `Tensor` of shape `[num_classes]`. The (possibly-partitioned)
class biases.
labels: A `Tensor` of type `int64` and shape `[batch_size,
num_true]`. The target classes. Note that this format differs from
the `labels` argument of `nn.softmax_cross_entropy_with_logits`.
inputs: A `Tensor` of shape `[batch_size, dim]`. The forward
activations of the input network.
num_sampled: An `int`. The number of classes to randomly sample per batch.
num_classes: An `int`. The number of possible classes.
num_true: An `int`. The number of target classes per training example.
sampled_values: a tuple of (`sampled_candidates`, `true_expected_count`,
`sampled_expected_count`) returned by a `*_candidate_sampler` function.
(if None, we default to `log_uniform_candidate_sampler`)
subtract_log_q: A `bool`. whether to subtract the log expected count of
the labels in the sample to get the logits of the true labels.
Default is True. Turn off for Negative Sampling.
remove_accidental_hits: A `bool`. whether to remove "accidental hits"
where a sampled class equals one of the target classes. Default is
False.
partition_strategy: A string specifying the partitioning strategy, relevant
if `len(weights) > 1`. Currently `"div"` and `"mod"` are supported.
Default is `"mod"`. See `tf.nn.embedding_lookup` for more details.
name: A name for the operation (optional).
seed: random seed for candidate sampling. Default to None, which doesn't set
the op-level random seed for candidate sampling.
Returns:
out_logits: `Tensor` object with shape
`[batch_size, num_true + num_sampled]`, for passing to either
`nn.sigmoid_cross_entropy_with_logits` (NCE) or
`nn.softmax_cross_entropy_with_logits` (sampled softmax).
out_labels: A Tensor object with the same shape as `out_logits`.
"""
if isinstance(weights, variables.PartitionedVariable):
weights = list(weights)
if not isinstance(weights, list):
weights = [weights]
with ops.name_scope(name, "compute_sampled_logits",
weights + [biases, inputs, labels]):
if labels.dtype != dtypes.int64:
labels = math_ops.cast(labels, dtypes.int64)
labels_flat = array_ops.reshape(labels, [-1])
# Sample the negative labels.
# sampled shape: [num_sampled] tensor
# true_expected_count shape = [batch_size, 1] tensor
# sampled_expected_count shape = [num_sampled] tensor
if sampled_values is None:
sampled_values = candidate_sampling_ops.log_uniform_candidate_sampler(
true_classes=labels,
num_true=num_true,
num_sampled=num_sampled,
unique=True,
range_max=num_classes,
seed=seed)
# NOTE: pylint cannot tell that 'sampled_values' is a sequence
# pylint: disable=unpacking-non-sequence
sampled, true_expected_count, sampled_expected_count = (
array_ops.stop_gradient(s) for s in sampled_values)
# pylint: enable=unpacking-non-sequence
sampled = math_ops.cast(sampled, dtypes.int64)
# labels_flat is a [batch_size * num_true] tensor
# sampled is a [num_sampled] int tensor
all_ids = array_ops.concat([labels_flat, sampled], 0)
# Retrieve the true weights and the logits of the sampled weights.
# weights shape is [num_classes, dim]
all_w = embedding_ops.embedding_lookup(
weights, all_ids, partition_strategy=partition_strategy)
if all_w.dtype != inputs.dtype:
all_w = math_ops.cast(all_w, inputs.dtype)
# true_w shape is [batch_size * num_true, dim]
true_w = array_ops.slice(all_w, [0, 0],
array_ops.stack(
[array_ops.shape(labels_flat)[0], -1]))
sampled_w = array_ops.slice(
all_w, array_ops.stack([array_ops.shape(labels_flat)[0], 0]), [-1, -1])
# inputs has shape [batch_size, dim]
# sampled_w has shape [num_sampled, dim]
# Apply X*W', which yields [batch_size, num_sampled]
sampled_logits = math_ops.matmul(inputs, sampled_w, transpose_b=True)
# Retrieve the true and sampled biases, compute the true logits, and
# add the biases to the true and sampled logits.
all_b = embedding_ops.embedding_lookup(
biases, all_ids, partition_strategy=partition_strategy)
if all_b.dtype != inputs.dtype:
all_b = math_ops.cast(all_b, inputs.dtype)
# true_b is a [batch_size * num_true] tensor
# sampled_b is a [num_sampled] float tensor
true_b = array_ops.slice(all_b, [0], array_ops.shape(labels_flat))
sampled_b = array_ops.slice(all_b, array_ops.shape(labels_flat), [-1])
# inputs shape is [batch_size, dim]
# true_w shape is [batch_size * num_true, dim]
# row_wise_dots is [batch_size, num_true, dim]
dim = array_ops.shape(true_w)[1:2]
new_true_w_shape = array_ops.concat([[-1, num_true], dim], 0)
row_wise_dots = math_ops.multiply(
array_ops.expand_dims(inputs, 1),
array_ops.reshape(true_w, new_true_w_shape))
# We want the row-wise dot plus biases which yields a
# [batch_size, num_true] tensor of true_logits.
dots_as_matrix = array_ops.reshape(row_wise_dots,
array_ops.concat([[-1], dim], 0))
true_logits = array_ops.reshape(_sum_rows(dots_as_matrix), [-1, num_true])
true_b = array_ops.reshape(true_b, [-1, num_true])
true_logits += true_b
sampled_logits += sampled_b
if remove_accidental_hits:
acc_hits = candidate_sampling_ops.compute_accidental_hits(
labels, sampled, num_true=num_true)
acc_indices, acc_ids, acc_weights = acc_hits
# This is how SparseToDense expects the indices.
acc_indices_2d = array_ops.reshape(acc_indices, [-1, 1])
acc_ids_2d_int32 = array_ops.reshape(
math_ops.cast(acc_ids, dtypes.int32), [-1, 1])
sparse_indices = array_ops.concat([acc_indices_2d, acc_ids_2d_int32], 1,
"sparse_indices")
# Create sampled_logits_shape = [batch_size, num_sampled]
sampled_logits_shape = array_ops.concat(
[array_ops.shape(labels)[:1],
array_ops.expand_dims(num_sampled, 0)], 0)
if sampled_logits.dtype != acc_weights.dtype:
acc_weights = math_ops.cast(acc_weights, sampled_logits.dtype)
sampled_logits += gen_sparse_ops.sparse_to_dense(
sparse_indices,
sampled_logits_shape,
acc_weights,
default_value=0.0,
validate_indices=False)
if subtract_log_q:
# Subtract log of Q(l), prior probability that l appears in sampled.
true_logits -= math_ops.log(true_expected_count)
sampled_logits -= math_ops.log(sampled_expected_count)
# Construct output logits and labels. The true labels/logits start at col 0.
out_logits = array_ops.concat([true_logits, sampled_logits], 1)
# true_logits is a float tensor, ones_like(true_logits) is a float
# tensor of ones. We then divide by num_true to ensure the per-example
# labels sum to 1.0, i.e. form a proper probability distribution.
out_labels = array_ops.concat([
array_ops.ones_like(true_logits) / num_true,
array_ops.zeros_like(sampled_logits)
], 1)
return out_logits, out_labels
其实这个函数就做了这么几件事:
- 采样:sampled_values如果为None,就是使用默认的采样方式 log_uniform_candidate_sampler。这个采样要求label list是按照频率降序排序的。
- 将positive labels和 negtive labels拼接起来,根据embedding_lookup去查找class labels的embedding表示;其实这个地方的weights和bias咱们是可以自己修改的,不一定是一组参数,可以是从模型得到的embedding。
- 根据label embedding和input进行matmul,得到logits,分别得到sample_logits和input_logits。注:其实input_logits的计算不是用matmul计算的,但是和matmul计算的结果是一样的。
- 去重。对采样样本正好是target label的样本的logits进行修改,由remove_accidental_hits变量标识是否去重。
- 减去log(Q)。
Differences between “sampled_softmax_loss” and “nce_loss”
其实两个loss的核心代码都是_compute_sampled_logits,但是在实现上不同的地方有两点:
- sampled_softmax_loss是有去重的,也就是remove_accidental_hits=True,但是nce_loss是不去重的,我们从上面的表也能看出来。
- sampled_softmax_loss采用的是softmax+CE,但是nce_loss采用的是sigmod+CE
再来看看两者实现:
sampled_softmax_loss
def sampled_softmax_loss(weights,
biases,
labels,
inputs,
num_sampled,
num_classes,
num_true=1,
sampled_values=None,
remove_accidental_hits=True,#去重
partition_strategy="mod",
name="sampled_softmax_loss",
seed=None):
"""Computes and returns the sampled softmax training loss.
This is a faster way to train a softmax classifier over a huge number of
classes.
This operation is for training only. It is generally an underestimate of
the full softmax loss.
A common use case is to use this method for training, and calculate the full
softmax loss for evaluation or inference. In this case, you must set
`partition_strategy="div"` for the two losses to be consistent, as in the
following example:
```python
if mode == "train":
loss = tf.nn.sampled_softmax_loss(
weights=weights,
biases=biases,
labels=labels,
inputs=inputs,
...,
partition_strategy="div")
elif mode == "eval":
logits = tf.matmul(inputs, tf.transpose(weights))
logits = tf.nn.bias_add(logits, biases)
labels_one_hot = tf.one_hot(labels, n_classes)
loss = tf.nn.softmax_cross_entropy_with_logits(
labels=labels_one_hot,
logits=logits)
```
See our [Candidate Sampling Algorithms Reference]
(https://www.tensorflow.org/extras/candidate_sampling.pdf)
Also see Section 3 of [Jean et al., 2014](http://arxiv.org/abs/1412.2007)
([pdf](http://arxiv.org/pdf/1412.2007.pdf)) for the math.
Args:
weights: A `Tensor` of shape `[num_classes, dim]`, or a list of `Tensor`
objects whose concatenation along dimension 0 has shape
[num_classes, dim]. The (possibly-sharded) class embeddings.
biases: A `Tensor` of shape `[num_classes]`. The class biases.
labels: A `Tensor` of type `int64` and shape `[batch_size,
num_true]`. The target classes. Note that this format differs from
the `labels` argument of `nn.softmax_cross_entropy_with_logits`.
inputs: A `Tensor` of shape `[batch_size, dim]`. The forward
activations of the input network.
num_sampled: An `int`. The number of classes to randomly sample per batch.
num_classes: An `int`. The number of possible classes.
num_true: An `int`. The number of target classes per training example.
sampled_values: a tuple of (`sampled_candidates`, `true_expected_count`,
`sampled_expected_count`) returned by a `*_candidate_sampler` function.
(if None, we default to `log_uniform_candidate_sampler`)
remove_accidental_hits: A `bool`. whether to remove "accidental hits"
where a sampled class equals one of the target classes. Default is
True.
partition_strategy: A string specifying the partitioning strategy, relevant
if `len(weights) > 1`. Currently `"div"` and `"mod"` are supported.
Default is `"mod"`. See `tf.nn.embedding_lookup` for more details.
name: A name for the operation (optional).
seed: random seed for candidate sampling. Default to None, which doesn't set
the op-level random seed for candidate sampling.
Returns:
A `batch_size` 1-D tensor of per-example sampled softmax losses.
"""
logits, labels = _compute_sampled_logits(
weights=weights,
biases=biases,
labels=labels,
inputs=inputs,
num_sampled=num_sampled,
num_classes=num_classes,
num_true=num_true,
sampled_values=sampled_values,
subtract_log_q=True,
remove_accidental_hits=remove_accidental_hits,
partition_strategy=partition_strategy,
name=name,
seed=seed)
labels = array_ops.stop_gradient(labels, name="labels_stop_gradient")
sampled_losses = nn_ops.softmax_cross_entropy_with_logits_v2(
labels=labels, logits=logits)
# sampled_losses is a [batch_size] tensor.
return sampled_losses
nce_loss
def nce_loss(weights,
biases,
labels,
inputs,
num_sampled,
num_classes,
num_true=1,
sampled_values=None,
remove_accidental_hits=False,#不去重
partition_strategy="mod",
name="nce_loss"):
"""Computes and returns the noise-contrastive estimation training loss.
See [Noise-contrastive estimation: A new estimation principle for
unnormalized statistical
models](http://www.jmlr.org/proceedings/papers/v9/gutmann10a/gutmann10a.pdf).
Also see our [Candidate Sampling Algorithms
Reference](https://www.tensorflow.org/extras/candidate_sampling.pdf)
A common use case is to use this method for training, and calculate the full
sigmoid loss for evaluation or inference. In this case, you must set
`partition_strategy="div"` for the two losses to be consistent, as in the
following example:
```python
if mode == "train":
loss = tf.nn.nce_loss(
weights=weights,
biases=biases,
labels=labels,
inputs=inputs,
...,
partition_strategy="div")
elif mode == "eval":
logits = tf.matmul(inputs, tf.transpose(weights))
logits = tf.nn.bias_add(logits, biases)
labels_one_hot = tf.one_hot(labels, n_classes)
loss = tf.nn.sigmoid_cross_entropy_with_logits(
labels=labels_one_hot,
logits=logits)
loss = tf.reduce_sum(loss, axis=1)
```
Note: By default this uses a log-uniform (Zipfian) distribution for sampling,
so your labels must be sorted in order of decreasing frequency to achieve
good results. For more details, see
`tf.random.log_uniform_candidate_sampler`.
Note: In the case where `num_true` > 1, we assign to each target class
the target probability 1 / `num_true` so that the target probabilities
sum to 1 per-example.
Note: It would be useful to allow a variable number of target classes per
example. We hope to provide this functionality in a future release.
For now, if you have a variable number of target classes, you can pad them
out to a constant number by either repeating them or by padding
with an otherwise unused class.
Args:
weights: A `Tensor` of shape `[num_classes, dim]`, or a list of `Tensor`
objects whose concatenation along dimension 0 has shape
[num_classes, dim]. The (possibly-partitioned) class embeddings.
biases: A `Tensor` of shape `[num_classes]`. The class biases.
labels: A `Tensor` of type `int64` and shape `[batch_size,
num_true]`. The target classes.
inputs: A `Tensor` of shape `[batch_size, dim]`. The forward
activations of the input network.
num_sampled: An `int`. The number of negative classes to randomly sample
per batch. This single sample of negative classes is evaluated for each
element in the batch.
num_classes: An `int`. The number of possible classes.
num_true: An `int`. The number of target classes per training example.
sampled_values: a tuple of (`sampled_candidates`, `true_expected_count`,
`sampled_expected_count`) returned by a `*_candidate_sampler` function.
(if None, we default to `log_uniform_candidate_sampler`)
remove_accidental_hits: A `bool`. Whether to remove "accidental hits"
where a sampled class equals one of the target classes. If set to
`True`, this is a "Sampled Logistic" loss instead of NCE, and we are
learning to generate log-odds instead of log probabilities. See
our [Candidate Sampling Algorithms Reference]
(https://www.tensorflow.org/extras/candidate_sampling.pdf).
Default is False.
partition_strategy: A string specifying the partitioning strategy, relevant
if `len(weights) > 1`. Currently `"div"` and `"mod"` are supported.
Default is `"mod"`. See `tf.nn.embedding_lookup` for more details.
name: A name for the operation (optional).
Returns:
A `batch_size` 1-D tensor of per-example NCE losses.
"""
logits, labels = _compute_sampled_logits(
weights=weights,
biases=biases,
labels=labels,
inputs=inputs,
num_sampled=num_sampled,
num_classes=num_classes,
num_true=num_true,
sampled_values=sampled_values,
subtract_log_q=True,
remove_accidental_hits=remove_accidental_hits,
partition_strategy=partition_strategy,
name=name)
sampled_losses = sigmoid_cross_entropy_with_logits(
labels=labels, logits=logits, name="sampled_losses")
# sampled_losses is batch_size x {true_loss, sampled_losses...}
# We sum out true and sampled losses.
return _sum_rows(sampled_losses)